6. Declustering and trend analysis
| 6.1 Distance betwen samples (download) | |
|
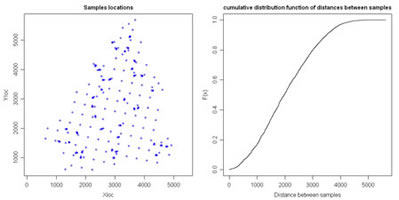
Compute distances between all pairs of samples and plot distances in a cumulative histogram. Variables to customize: varcolXY <- c(1,2) # index of the GTD frame X and Y coordinatessizePOINTS <- 1 # size of the sample symbols of the left plot |
 |
| 6.2 Compute Vononoi Polygons (download) | |
|
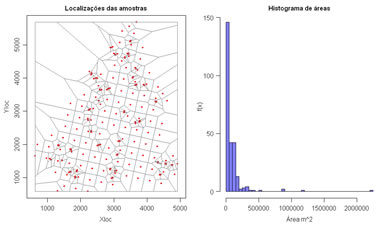
Compute Voronoi polygons and compute declustered basic statistics. An histogram of the Voronoi polygon areas are displayed at right. Variables to customize: varcolXY <- c(1,2) # index of the GTD frame X and Y coordinates |
 |
| 6.3 Cell declustering (download) | |
|
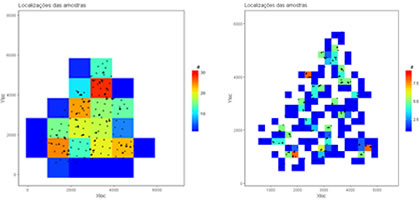
Overlap a set of samples with a mesh of cells and compute average values of several variables within each cell. After, compute global declustered basic statistics. Variables to customize: varcolXY <- c(1,2) # index of the GTD frame X and Y coordinates |
 |
| 6.4 Spatial trend analysis in X and Y directions (download) | |
|
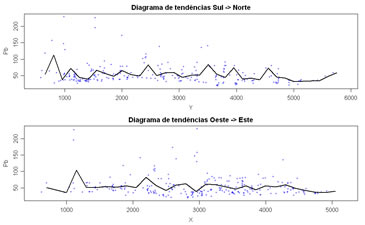
Perform a trend analysis of a quantitative variable along X and Y directions. Variables to customize: varcolXY <- c(1,2) # index of the GTD frame X and Y coordinatesvarlistindex <- 10 # index of the GTD quantitative variable to trend analysis |
 |
