5. Multivariate data analysis
| 5.1 Standardization of quantitative variables (download) | |
|
Apply a standardization formula for a set of quantitative variables. Three types of standardization can be donne: Variables to customize: varlistindex <- c(5,6,7,8,9,10,11) # list of variables of GTD frame to standardizetipo <- 1 # standardization type (1), (2) or (3)ndecimals <- 3 # number of decimals |
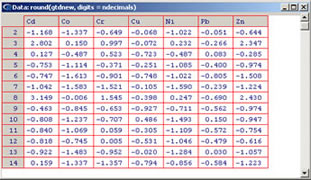 |
| 5.2 Principal Component Analysis - PCA (download) | |
|
Execute a PCA for a set of quantitative variables. Four graphics are displayed as results: (1) eigenvalues histogram; (2) graphic of correlations between the quantitative variables and the Principal Components; (3) another graphic of correlations between the quantitative variables and pairs of Principal Components; (4) projection of the individuals in the Principal Components. Variables to customize: varlistindex <- c(5,6,7,8,9,10,11) # index of the GTD frame Qualitative Variables to execute PCA |
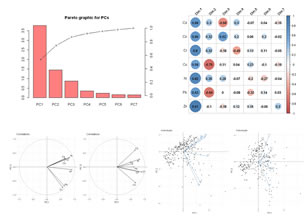 |
| 5.3 Principal Component Analysis step by syep (download) | |
|
Execute a PCA step-by-step for a set of quantitative variables, in a way that all details and matrices can be followed and listed. Variables to customize: varlistindex <- c(5,6,7,8,9,10,11) # index of the GTD frame Qualitative Variables to execute PCA |
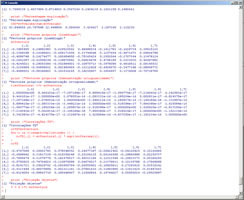 |
| 5.4 Hierarchical clustering (download) | |
|
Perform a hierarchical clustering of a set of quantitative variables. Distance type and aglomeration mode should be selected. A specific number of groups can be created and conditional basic statistica for each group are diaplayed. Created groups can be seen on PCA representations. Variables to customize: varlistindex <- c(5,6,7,8,9,10,11) # index of the GTD quantitative variables to execute a Hierarchical Clustering |
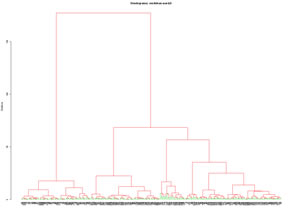 |
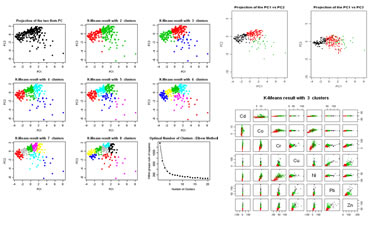
| 5.5 K-means clustering (download) | |
|
Clustering sets of individuals by using the K-means algorithm. This algorithm can run in two modes: (1) evaluation mode, trial for creation of several groups and evaluation of results; (2) separate the data into a number of previously defined groups. For (2) classification can be made with original data or the two first Principal Components. Created groups can be seen on PCA repreentations. Variables to customize: varlistindex <- c(5,6,7,8,9,10,11) # index of the GTD quantitative variables to execute a K-means |
 |
